RStudio is an integrated development environment (IDE) for R. It includes a console, syntax-highlighting editor that supports direct code execution, as well as tools for plotting, history, debugging and workspace management.
Publish to RPubs
Getting Started with RPubs
RStudio lets you harness the power of R Markdown to create documents that weave together your writing and the output of your R code. And now, with RPubs, you can publish those documents on the web with the click of a button!
Prerequisites
You’ll need R itself, RStudio (v0.96.230 or later), and the knitr package (v0.5 or later).
Instructions
In RStudio, create a new R Markdown document by choosing File | New | R Markdown.
Click the Knit HTML button in the doc toolbar to preview your document.
In the preview window, click the Publish button.
Results:
Convergence testing1
2
3
4
5
6
7
8library(lattice)
library(lme4)
library(blme)
library(reshape2)
library(ggplot2); theme_set(theme_bw())
library(gridExtra) ## for grid.arrange
library(bbmle) ## for slice2D; requires *latest* r-forge version (r121)
source("allFit.R")
Load data:1
dList <- load("data.RData")
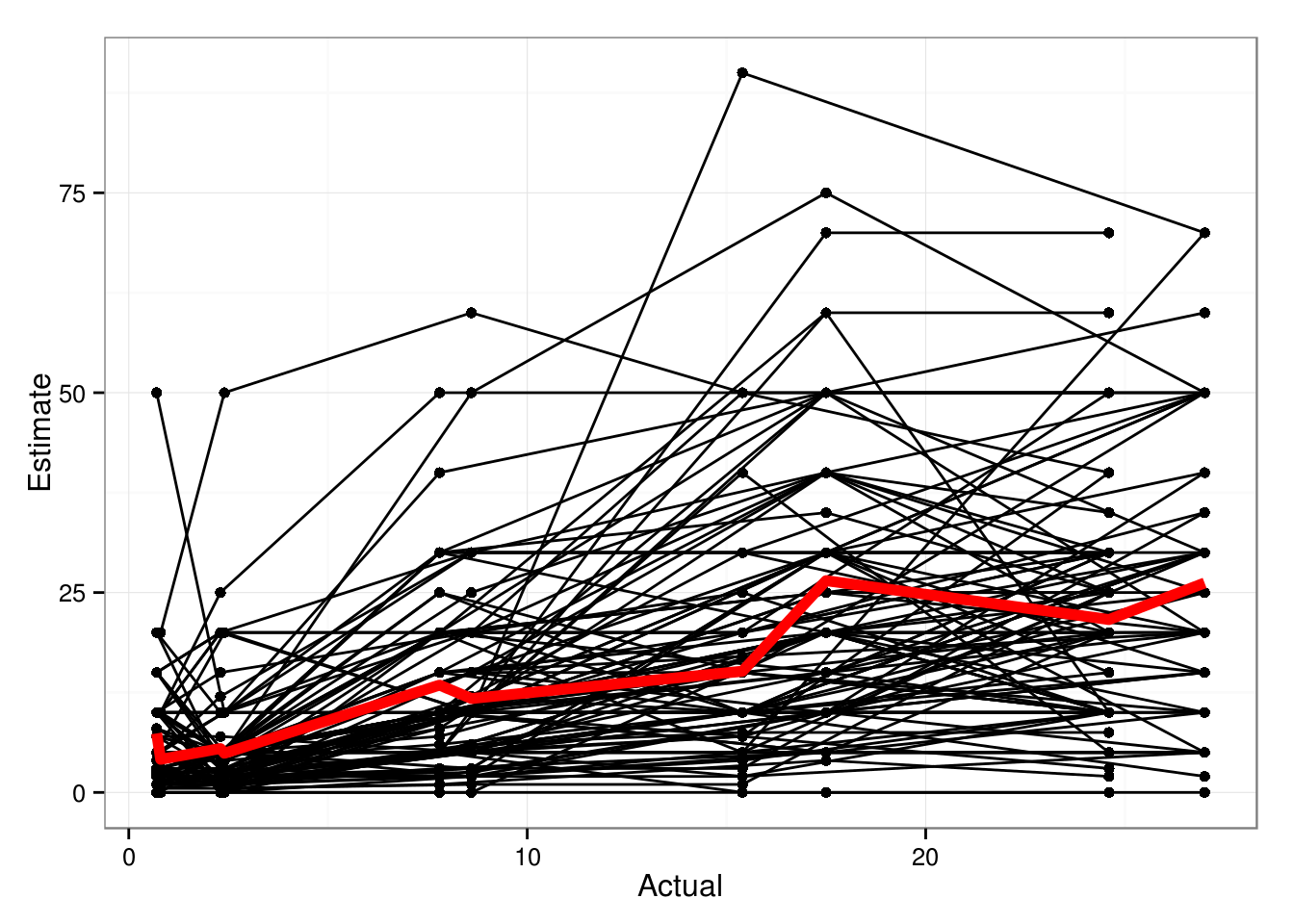
Spaghetti plot: don’t see much pattern other than (1) general increasing trend; (2) quantized response values (table(dth$Estimate) or unique(dth$Estimate) also show this); (3) skewed residuals1
sort(unique(dth$Estimate))
1 | ## [1] 0.0 0.5 1.0 1.5 2.0 2.5 3.0 4.0 5.0 6.0 7.0 7.5 8.0 10.0 |
1 | (p0 <- ggplot(dth,aes(Actual,Estimate))+geom_point()+ |