1. FastQC察看
2. 进行reads的修剪和过滤
Short-insert paired end reads
接头序列:1
2
3
4PrefixPE/1
TACACTCTTTCCCTACACGACGCTCTTCCGATCT
PrefixPE/2
GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT
Trimmomatic等通常的质控软件。
Long Mate Pair libraries
接头序列:technote_nextera_matepair_data_processing.pdf
针对此类数据的处理软件主要是:nextclip和skewer,从文章结果来看后者略优。【Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads】
处理软件:nextclip (同时移除PCR duplicates)
1 | ./nextclip -d -i ~/AS/raw_reads/AS8K_R1.fastq -j ~/AS/raw_reads/AS8K_R2.fastq -o output |
如上,若出现too much rehashing!! Rehash=26的错误信息则增大[-n | --number_of_reads] Approximate number of reads (default 20,000,000)参数值;1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124./nextclip -d -e -i ~/AS/raw_reads/AS3K_R1.fastq -j ~/AS/raw_reads/AS3K_R2.fastq -o 33AS3K -n 30000000
NextClip v1.3.2
n: 19
b: 100
Entries: 52428800
Entry size: 24
Memory required: 1456 MB
Creating hash tables for duplicate storage...
Hash:
unique kmers: 0
Capacity: 52428800
Occupied: 0.00%
Pruned: 0 (-nan%)
Collisions:
Adaptor: CTGTCTCTTATACACATCTAGATGTGTATAAGAGACAG
Opening input filename /public/home/zpxu/AS/raw_reads/AS3K_R1.fastq
Opening input filename /public/home/zpxu/AS/raw_reads/AS3K_R2.fastq
Opening output file 33AS3K_A_R1.fastq
Opening output file 33AS3K_A_R2.fastq
Opening output file 33AS3K_B_R1.fastq
Opening output file 33AS3K_B_R2.fastq
Opening output file 33AS3K_C_R1.fastq
Opening output file 33AS3K_C_R2.fastq
Opening output file 33AS3K_D_R1.fastq
Opening output file 33AS3K_D_R2.fastq
Opening output file 33AS3K_E_R1.fastq
Opening output file 33AS3K_E_R2.fastq
Warning: read shorter than minimum read size (64) - ignoring
GC bases: 9705583313 AT bases: 12168077887
Hash:
unique kmers: 28281698
Capacity: 52428800
Occupied: 53.94%
Pruned: 0 (0.00%)
Collisions:
tries 0: 72912204
Counting duplicates...
8% [======== ]Warning: count (999) exceeds maximum - treated as 999
10% [========== ]Warning: count (999) exceeds maximum - treated as 999
20% [==================== ]Warning: count (999) exceeds maximum - treated as 999
23% [======================= ]Warning: count (999) exceeds maximum - treated as 999
25% [========================= ]Warning: count (999) exceeds maximum - treated as 999
67% [=================================================================== ]Warning: count (999) exceeds maximum - treated as 999
83% [=================================================================================== ]Warning: count (999) exceeds maximum - treated as 999
100% [====================================================================================================]
SUMMARY
Strict match parameters: 34, 18
Relaxed match parameters: 32, 17
Minimum read size: 25
Trim ends: 19
Number of read pairs: 72966745
Number of duplicate pairs: 44626706 61.16 %
Number of pairs containing N: 54541 0.07 %
R1 Num reads with adaptor: 16173479 22.17 %
R1 Num with external also: 4375021 6.00 %
R1 long adaptor reads: 11092453 15.20 %
R1 reads too short: 5081026 6.96 %
R1 Num reads no adaptor: 12162760 16.67 %
R1 no adaptor but external: 5248876 7.19 %
R2 Num reads with adaptor: 14902833 20.42 %
R2 Num with external also: 4406543 6.04 %
R2 long adaptor reads: 9987006 13.69 %
R2 reads too short: 4915827 6.74 %
R2 Num reads no adaptor: 13433406 18.41 %
R2 no adaptor but external: 5653578 7.75 %
Total pairs in category A: 11389962 15.61 %
A pairs long enough: 5627734 7.71 %
A pairs too short: 5762228 7.90 %
A external clip in 1 or both: 18225 0.02 %
A bases before clipping: 3416988600
A total bases written: 749338798
Total pairs in category B: 3422082 4.69 %
B pairs long enough: 1695273 2.32 %
B pairs too short: 1726809 2.37 %
B external clip in 1 or both: 47947 0.07 %
B bases before clipping: 1026624600
B total bases written: 323696037
Total pairs in category C: 4565902 6.26 %
C pairs long enough: 2610991 3.58 %
C pairs too short: 1954911 2.68 %
C external clip in 1 or both: 143843 0.20 %
C bases before clipping: 1369770600
C total bases written: 509149505
Total pairs in category D: 8649889 11.85 %
D pairs long enough: 3667738 5.03 %
D pairs too short: 4982151 6.83 %
D external clip in 1 or both: 5627647 7.71 %
D bases before clipping: 2594966700
D total bases written: 899148840
Total pairs in category E: 308404 0.42 %
E pairs long enough: 196969 0.27 %
E pairs too short: 111435 0.15 %
E external clip in 1 or both: 37111 0.05 %
E bases before clipping: 92521200
E total bases written: 29751268
Total usable pairs: 10130967 13.88 %
All long enough: 13798705 18.91 %
All categories too short: 14537534 19.92 %
Duplicates not written: 44630506 61.17 %
Category B became E: 90789 0.12 %
Category C became E: 217615 0.30 %
Overall GC content: 44.37 %
Done. Completed in 4414 seconds.
结果文件中的A,B和C category合并后用于后续分析。
处理软件: skewer
1 | ./skewer -m mp -i ~/AS/raw_reads/AS8K_R1.fastq ~/AS/raw_reads/AS8K_R2.fastq -o AS8K -t 5 |
两类reads的去除比例
After trimming and quality filtering, 56% of long-insert reads from each of the three mate-pair libraries and 95% of paired-end reads were retained on average.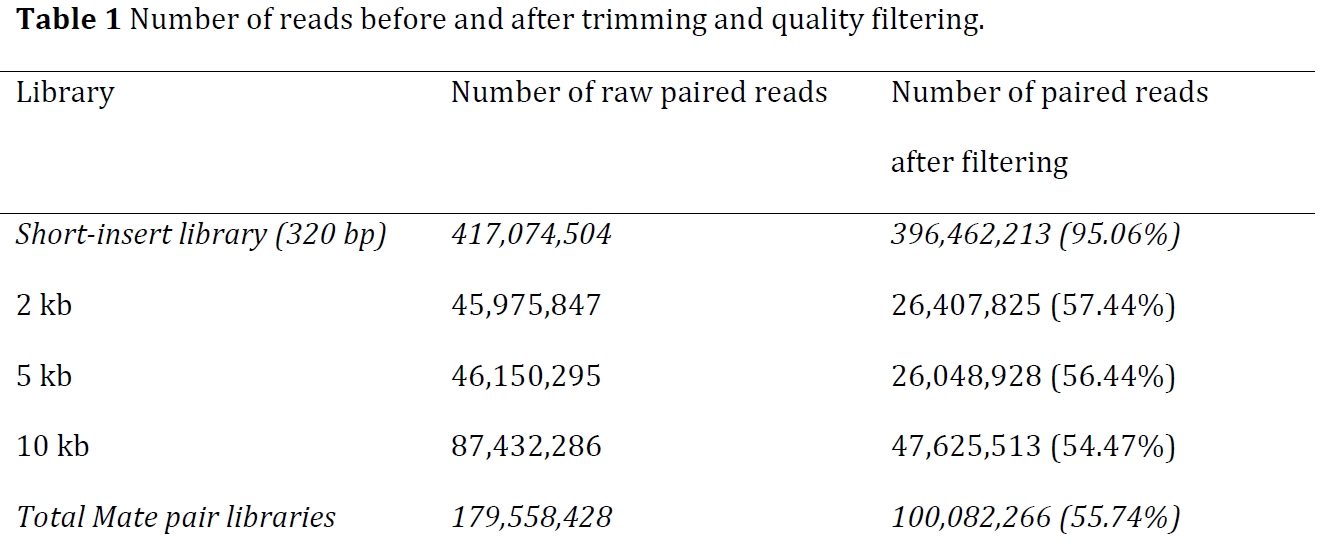
实际同样数据运行结果比较
| FastQ files | read length | median | mean | stdev | FF | FR | RF | RR |
|---|---|---|---|---|---|---|---|---|
| AS3K_R2_nextclip.fq.is.txt | 83 | 2510 | 2307.246 | 764.2089 | 36 | 781 | 9164 | 19 |
| AS5K_R2_nextclip.fq.is.txt | 84 | 4444 | 3974.787 | 1506.76 | 54 | 773 | 9127 | 46 |
| AS8K_R2_nextclip.fq.is.txt | 81 | 5825 | 4733.265 | 2530.765 | 150 | 1167 | 8543 | 140 |
| AS3K-trimmed-pair2.fastq.is.txt.skewer | 102 | 2493 | 2319.141 | 725.1056 | 26 | 1912 | 8049 | 13 |
| AS5K-trimmed-pair2.fastq.is.txt.skewer | 106 | 4460 | 4042.562 | 1443.632 | 30 | 2333 | 7608 | 29 |
| AS8K-trimmed-pair2.fastq.is.txt.skewer | 111 | 5945 | 4935.393 | 2446.029 | 112 | 3607 | 6212 | 69 |
3. FastUniq 去除 paired reads 的PCR重复
建议先trim,然后在来用这个软件来去除dup,因为,这个软件是比较以后,随机保留相同的pair的中一个,如果不先trim,容易保留质量差的哪一个,而且即使trim后,它也能处理不同长度的pair。 【每日一生信—FastUniq去除paired reads的duplicates】
单个文库/input_list.txt 多次运行
1 | cat AS285.list |
或者多个文库写在同一个input_list.txt时输出结果会将多个文库合并成一个文件;
1 | cat input_list.txt |
报错:内存问题,在大内存节点运行。1
Error in Reading pair-end FASTQ sequence!
4. 进行reads 的纠正
BLESS和Musket有相似的纠正结果,前者一直报错;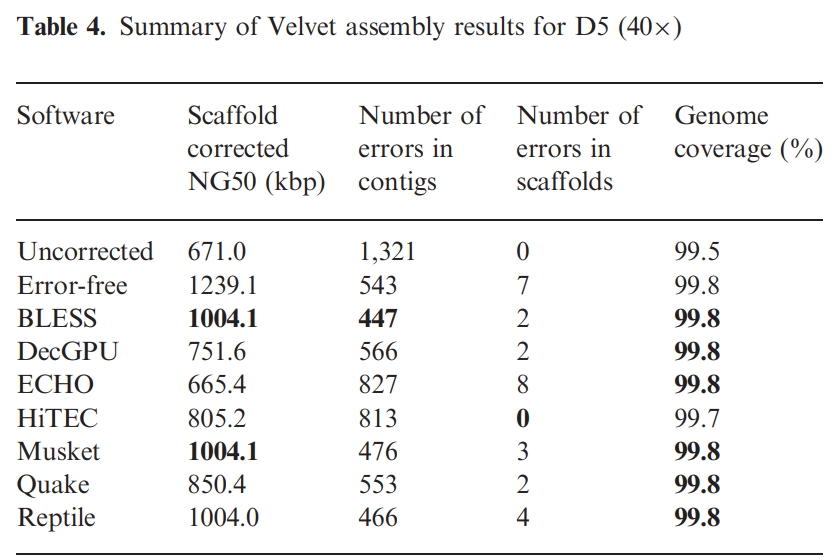
BLESS
1 | source /public/home/software/.bashrc |
报错:1
2
3Checking input read files
ERROR: Irregular quality score range 35-75
Musket - a multistage k-mer spectrum based corrector
1 | musket AS485_R1.rd.clean.fastq AS485_R2.rd.clean.fastq -omulti AS485 -inorder -p 10 |
至此,经过Trim,去PCR duplicates和纠正后的reads可用于后续的基因组组装等其他分析。