命令行访问和获取NCBI数据当选Entrez Direct: E-utilities on the UNIX Command Line.
工具集
esearch 搜索功能;
elink looks up neighbors (within a database) or links (between databases).
efilter 搜索结果过滤,搜索结果以特定格式输出.
efetch 以指定格式下载搜索结果.
xtract 转化XML格式为table.
einfo obtains information on indexed fields in an Entrez database.
epost uploads unique identifiers (UIDs) or sequence accession numbers.
nquire sends a URL request to a web page or CGI service.
数据库查询
1 | esearch -db pubmed -query "lycopene cyclase" | efetch -format abstract |
当查询数据是蛋白或核酸时-format参数可以是fasta(fasta_cds_na, fasta_cds_aa, and gene_fasta),gb(GenBank), gp(GenPept),
搜索和过滤
1 | esearch -db pubmed -query "opsin gene conversion" | elink -related | |
场景需求:linux下如何完成如下检索和筛选过程?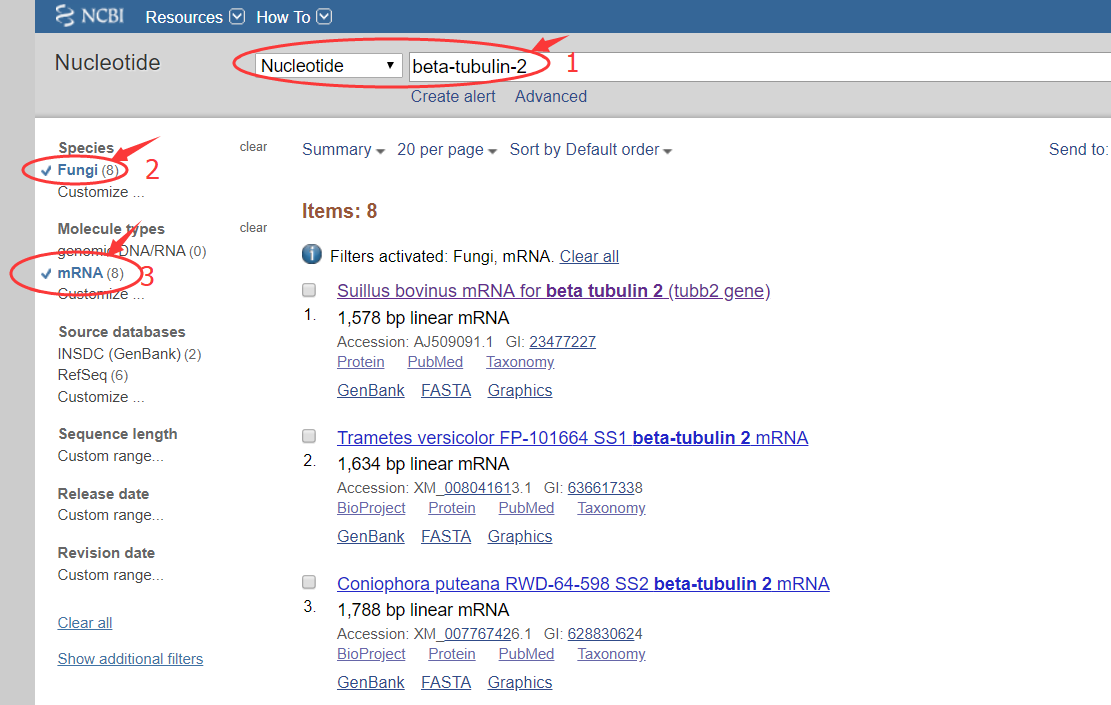
在第一步query时添加筛选项:1
esearch -db nucleotide -query "beta-tubulin-2 AND (Fungi[filter] AND "mrna"[Filter])" | efetch -format fasta
XML格式转换为制表符
1 | $ esearch -db protein -query "lycopene cyclase" |
某一领域最多产的作者
1 | $ SortUniqCountRank() { |
某一领域每年文章发表情况
1 | esearch -db pubmed -query "legionnaires disease [TITL]" | |
人每条染色体上有多少基因
1 | for chr in {1..22} X Y MT |
