Degradome sequencing
Degradome sequencing (Degradome-Seq),also referred to as parallel analysis of RNA ends (PARE),is a modified version of 5’-Rapid Amplification of cDNA Ends (RACE) using high-throughput, deep sequencing method using as Illumina’s SBS technology. Degradome sequencing provides a comprehensive means of analyzing patterns of RNA degradation.
Degradome sequencing has been used to identify microRNA (miRNA) cleavage sites,because miRNAs can cause endonucleolytic cleavage of mRNA by extensive and often perfect complementarity to mRNAs.Degradome sequencing revealed many known and novel plant miRNA (siRNA) targets. Recently, degradome sequencing also has been applied to identify animal (human and mouse) miRNA-derived cleavages.
原理
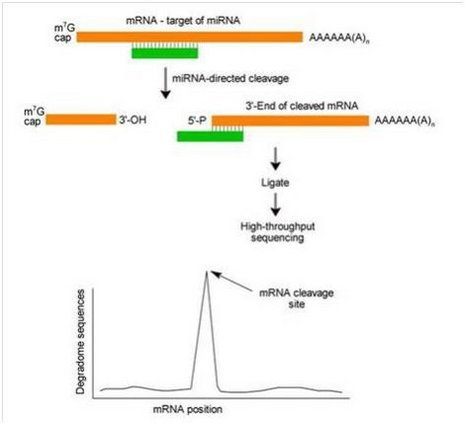
在植物体内绝大多数的miRNA是利用剪切作用调控靶基因的表达,且剪切常发生在miRNA与mRNA互补区域的第十位核苷酸上。靶基因经剪切产生二个片段,5’ 剪切片段和3’ 剪切片段。其中3’ 剪切片段,包含有自由的5’ 单磷酸和3’ polyA尾巴,可被RNA连接酶连接,连接产物可用于下游高通量测序;而含有5’ 帽子结构的完整基因,含有帽子结构的5’ 剪切片段或是其他缺少5’ 单磷酸基团的RNA是无法被RNA酶连接的,因而无法进入下游的测序实验;对测序数据进行深入地比对分析,可以直观地发现在mRNA序列的某个位点会出现一个波峰,而该处正是候选的miRNA剪切位点。
測序數據的分析方法
測序數據使用賓州大學 Addo-Quaye 等建立CleaveLand 分析方法進行比對分析,可以直觀地發現在 mRNA 序列的某個位點會出現一個波峰,而該處正是候選的 miRNA 剪切位元點。
