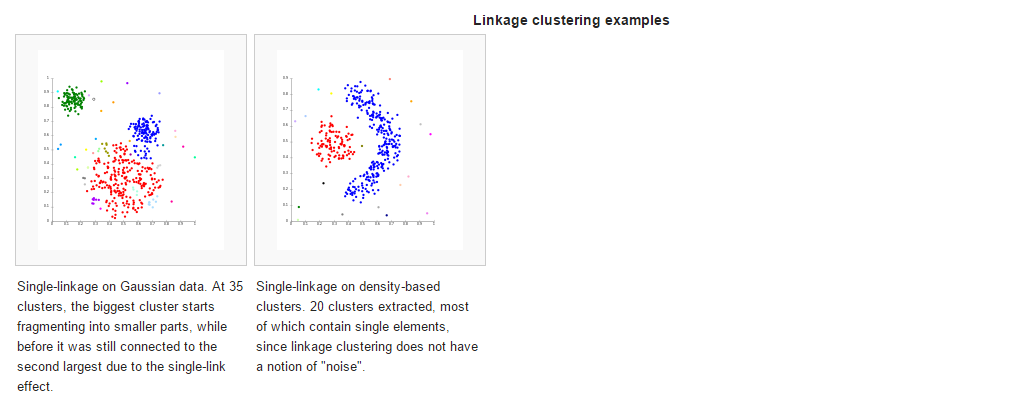
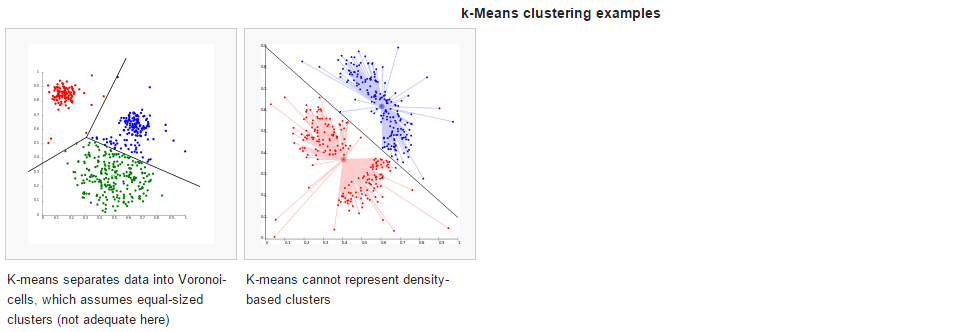
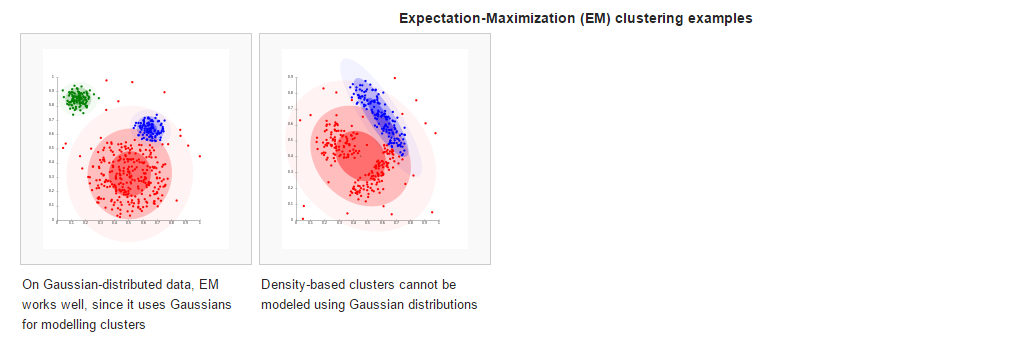
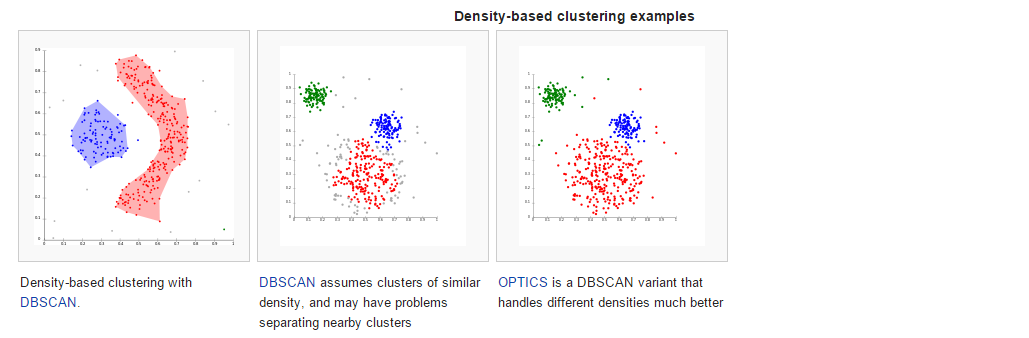
准备素材
1 | $ cat facebook.txt |
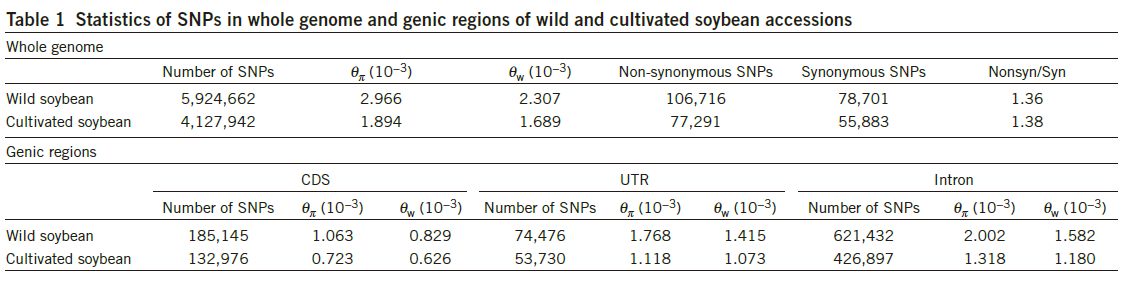
第一个域是公司名称,第二个域是公司人数,第三个域是员工平均工资。
排序
让这个文件按公司的字母顺序排序,也就是按第一个域进行排序:(这个facebook.txt文件有三个域)1
2
3
4
5$ sort -t ' ' -k 1 facebook.txt
baidu 100 5000
google 110 5000
guge 50 3000
sohu 100 4500
看到了吧,就直接用-k 1设定就可以了。(其实此处并不严格,稍后你就会知道)
若数据存在表头,而我们一般不需要对表头排序,但是又希望排序后表头依然存在,该如何操作呢?1
2## if you have two header lines and want to keep both of them:
(sed -n '1,2p' your_file; cat your_file | sed '1,2d' | sort) > sort_header.txt
让facebook.txt按照公司人数排序1
2
3
4
5$ sort -n -t ' ' -k 2 facebook.txt
guge 50 3000
baidu 100 5000
sohu 100 4500
google 110 5000
但是,此处出现了问题,那就是baidu和sohu的公司人数相同,都是100人,这个时候怎么办呢?按照默认规矩,是从第一个域开始进行升序排序,因此baidu排在了sohu前面。
让facebook.txt按照公司人数排序 ,人数相同的按照员工平均工资升序排序:1
2
3
4
5$ sort -n -t ' ' -k 2 -k 3 facebook.txt
guge 50 3000
sohu 100 4500
baidu 100 5000
google 110 5000
加了一个-k2 -k3就解决了问题,sort支持这种设定,就是说设定域排序的优先级,先以第2个域进行排序,如果相同,再以第3个域进行排序。(如果你愿意,可以一直这么写下去,设定很多个排序优先级)
让facebook.txt按照员工工资降序排序,如果员工人数相同的,则按照公司人数升序排序:1
2
3
4
5$ sort -n -t ' ' -k 3r -k 2 facebook.txt
baidu 100 5000
google 110 5000
sohu 100 4500
guge 50 3000
此处有使用了一些小技巧,你仔细看看,在-k 3后面偷偷加上了一个小写字母r,r和-r选项的作用是一样的,就是表示逆序。因为sort默认是按照升序排序的,所以此处需要加上r表示第三个域(员工平均工资)是按照降序排序。此处你还可以加上n,就表示对这个域进行排序时,要按照数值大小进行排序,举个例子吧:1
2
3
4
5$ sort -t ' ' -k 3nr -k 2n facebook.txt
baidu 100 5000
google 110 5000
sohu 100 4500
guge 50 3000
看,我们去掉了最前面的-n选项,而是将它加入到了每一个-k选项中了。
-k选项的具体语法格式
要继续往下深入的话,就不得不来点理论知识。你需要了解-k选项的语法格式,如下:1
[ FStart [ .CStart ] ] [ Modifier ] [ , [ FEnd [ .CEnd ] ][ Modifier ] ]
这个语法格式可以被其中的逗号(”,”)分为两大部分,Start部分和End部分。
先给你灌输一个思想,那就是”如果不设定End部分,那么就认为End被设定为行尾”。这个概念很重要的,但往往你不会重视它。
Start部分也由三部分组成,其中的Modifier部分就是我们之前说过的类似n和r的选项部分。我们重点说说Start部分的FStart和C.Start。
C.Start也是可以省略的,省略的话就表示从本域的开头部分开始。之前例子中的-k 2和-k 3就是省略了C.Start的例子喽。
FStart.CStart,其中FStart就是表示使用的域,而CStart则表示在FStart域中从第几个字符开始算”排序首字符”。
同理,在End部分中,你可以设定FEnd.CEnd,如果你省略.CEnd,则表示结尾到”域尾”,即本域的最后一个字符。或者,如果你将CEnd设定为0(零),也是表示结尾到”域尾”。
从公司英文名称的第二个字母开始进行排序:1
2
3
4
5$ sort -t ' ' -k 1.2 facebook.txt
baidu 100 5000
sohu 100 4500
google 110 5000
guge 50 3000
使用了-k 1.2,这就表示对第一个域的第二个字符开始到本域的最后一个字符为止的字符串进行排序。你会发现baidu因为第二个字母是a而名列榜首。sohu和google第二个字符都是o,但sohu的h在google的o前面,所以两者分别排在第二和第三。guge只能屈居第四了。
只针对公司英文名称的第二个字母进行排序,如果相同的按照员工工资进行降序排序:1
2
3
4
5$ sort -t ' ' -k 1.2,1.2 -k 3,3nr facebook.txt
baidu 100 5000
google 110 5000
sohu 100 4500
guge 50 3000
由于只对第二个字母进行排序,所以我们使用了-k 1.2,1.2的表示方式,表示我们”只”对第二个字母进行排序。(如果你问”我使用-k 1.2怎么不行?”,当然不行,因为你省略了End部分,这就意味着你将对从第二个字母起到本域最后一个字符为止的字符串进行排序)。对于员工工资进行排序,我们也使用了-k 3,3,这是最准确的表述,表示我们”只”对本域进行排序,因为如果你省略了后面的3,就变成了我们”对第3个域开始到最后一个域位置的内容进行排序”了。
在modifier部分还可以用到哪些选项?
可以用到b、d、f、i、n 或 r。
其中n和r你肯定已经很熟悉了。
b表示忽略本域的签到空白符号。
d表示对本域按照字典顺序排序(即,只考虑空白和字母)。
f表示对本域忽略大小写进行排序。
i表示忽略”不可打印字符”,只针对可打印字符进行排序。(有些ASCII就是不可打印字符,比如\a是报警,\b是退格,\n是换行,\r是回车等等)
思考关于-k和-u联合使用的例子:1
2
3
4
5$ cat facebook.txt
google 110 5000
baidu 100 5000
guge 50 3000
sohu 100 4500
这是最原始的facebook.txt文件。1
2
3
4
5
6
7
8
9$ sort -n -k 2 facebook.txt
guge 50 3000
baidu 100 5000
sohu 100 4500
google 110 5000
$ sort -n -k 2 -u facebook.txt
guge 50 3000
baidu 100 5000
google 110 5000
当设定以公司员工域进行数值排序,然后加-u后,sohu一行就被删除了!原来-u只识别用-k设定的域,发现相同,就将后续相同的行都删除。1
2
3
4
5
6
7
8
9
10$ sort -k 1 -u facebook.txt
baidu 100 5000
google 110 5000
guge 50 3000
sohu 100 4500
$ sort -k 1.1,1.1 -u facebook.txt
baidu 100 5000
google 110 5000
sohu 100 4500
这个例子也同理,开头字符是g的guge就没有幸免于难。1
2
3
4
5$ sort -n -k 2 -k 3 -u facebook.txt
guge 50 3000
sohu 100 4500
baidu 100 5000
google 110 5000
咦!这里设置了两层排序优先级的情况下,使用-u就没有删除任何行。原来-u是会权衡所有-k选项,将都相同的才会删除,只要其中有一级不同都不会轻易删除的:)(不信,你可以自己加一行sina 100 4500试试看)
最诡异的排序:1
2
3
4
5$ sort -n -k 2.2,3.1 facebook.txt
guge 50 3000
baidu 100 5000
sohu 100 4500
google 110 5000
以第二个域的第二个字符开始到第三个域的第一个字符结束的部分进行排序。
第一行,会提取0 3,第二行提取00 5,第三行提取00 4,第四行提取10 5。
又因为sort认为0小于00小于000小于0000….
因此0 3肯定是在第一个。10 5肯定是在最后一个。但为什么00 5却在00 4前面呢?(你可以自己做实验思考一下。)
答案揭晓:原来”跨域的设定是个假象”,sort只会比较第二个域的第二个字符到第二个域的最后一个字符的部分,而不会把第三个域的开头字符纳入比较范围。当发现00和00相同时,sort就会自动比较第一个域去了。当然baidu在sohu前面了。用一个范例即可证实:1
2
3
4
5$ sort -n -k 2.2,3.1 -k 1,1r facebook.txt
guge 50 3000
sohu 100 4500
baidu 100 5000
google 110 5000
有时候在sort命令后会看到+1 -2这些符号,这是什么东东?
关于这种语法,最新的sort是这么进行解释的:
On older systems, sort' supports an obsolete origin-zero syntax+POS1 [-POS2]’ for specifying sort keys. POSIX 1003.1-2001 (*note Standards conformance::) does not allow this; use `-k’ instead.
原来,这种古老的表示方式已经被淘汰了,以后可以理直气壮的鄙视使用这种表示方法的脚本喽!
(为了防止古老脚本的存在,在这再说一下这种表示方法,加号表示Start部分,减号表示End部分。最最重要的一点是,这种方式方法是从0开始计数的,以前所说的第一个域,在此被表示为第0个域。以前的第2个字符,在此表示为第1个字符.)
其他有用参数
V(大写):聪明的字母和数字排序
1 | cat example2.bed |
V is only for GNU sort which will sort chromsome in alpha-numeric order。
设置临时目录
1 | -T, --temporary-directory=DIR |
因为sort命令默认临时文件目录为”/tmp”,在sort执行过程中,如果产生的临时文件过大会导致”/tmp”目录被占满的。
Contribution from :
本原创文章属于《Linux大棚》博客,博客地址为http://roclinux.cn。文章作者为rocrocket。